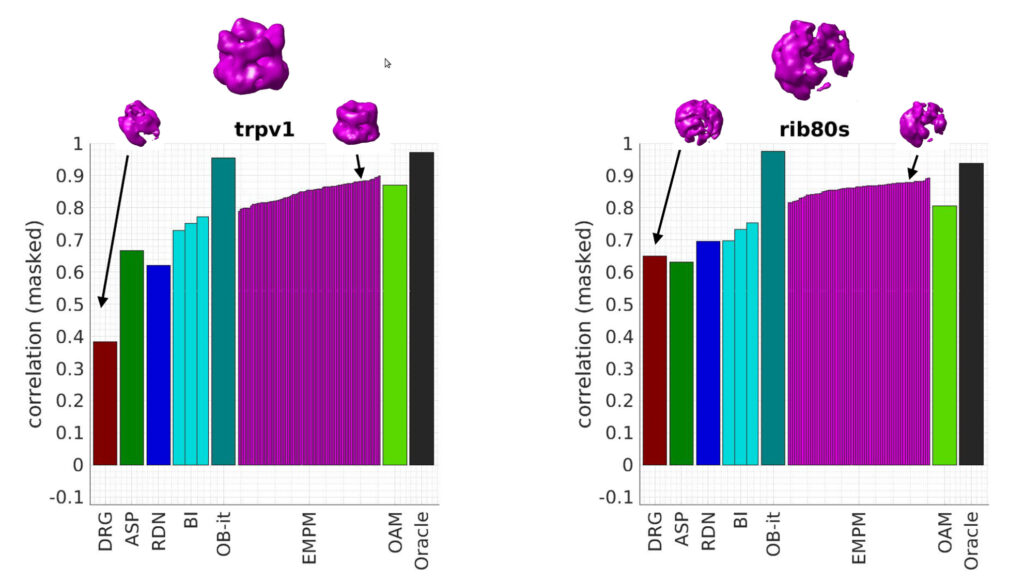
We are delighted to announce that we will be hosting a pilot virtual One World Cryo-EM Poster Session on March 10-11, 2021. This poster session, held on gather.town, will be a chance to discuss research with colleagues, share your own work, and socialize with others in the community.
A list of posters, presenters, and their availability can be found below or at the following link.
We have 3 live poster sessions on: March 10 11am-1pm EST, March 10 8-10pm EST, March 11 8am-10am EST.
If you are interested in presenting, please submit your poster on this application form. With your poster, you may optionally submit a 3-5 minute video to be displayed alongside your poster for visitors outside of live poster sessions. More information and details are provided in the form.
Please make sure that you are on our mailing list. This is the only place where the links to the gather.town poster session will be posted. As always, you can find more information at https://cryoem.world.
We are excited to host this poster session, and hope that you will join us.
Important Rules and Policies
As a participant, you agree that any information presented in the One World Cryo-EM poster session, whether in a poster, talk, video, or discussion, is private communication and that the information is not for public use.
You agree not to record the information by any means and not to quote or publish it without the written permission of the presenter. This includes posting information on social media, in publications, and on blogs.
You understand that the posters are not peer-reviewed and not archivable.
You understand that it is up to each participant to follow these restrictions. The organizers are not responsible for enforcing these restrictions.
Note that the list of poster titles and authors will be available on the OWCryo-EM website. Therefore, you may tweet a recommendation to visit poster #123 titled x by y (#OWCryoEMPosters) without referencing the content.
As a presenter, you may post your own poster on Twitter and invite people to retweet it and meet you at your poster #OWCryoEMPosters.
FAQ
Q: What is gather.town? What do posters there look like?
A: This is what a poster looks like on gather.town: https://gather.town/app/5Qb5FSmPM0wtDvpd/sample_poster (the actual session is expected to have a slightly larger number of posters).
Q: How do I view the gather.town session?
A: Please make sure that you are on our mailing list. This is the only place where the links to the gather.town poster session will be posted.
Q: What is the poster selection process?
A: In this pilot poster session, we expect to accept all novel work that satisfies the topic, quality, and form requirements.
Q: How do I submit a poster?
A: Please submit your completed poster on this application form. You are not required to submit a title and abstract before submitting the full poster.
Q: What are the requirements for the poster?
A: Posters should be in an image format (eg .png, .jpg, but not .pdf), and should be at least 4000×6000 pixels. You may resubmit posters up to 10 times until February 22nd. These will be collectively posted in gather.town, and we will let you know your poster ID number. Here is a poster size example.
Q: What are the requirements for the video?
A: Videos are optional, and should be between 3-5 minutes long and provide viewers a brief description of your poster. These will be posted next to your poster for visitors that cannot make the live session, or to provide visitors context about your poster.