Peter Schwander
PhD, Assoc. Professor
Date and time: June 8th, 2022
Wednesday, at 8am PT / 11am ET / 4pm BST / 5pm CEST / 11pm China
For the zoom and gather.town links, please join the One World Cryo-EM mailing list.
Routes to Extract Biological Function from single-particle cryoEM
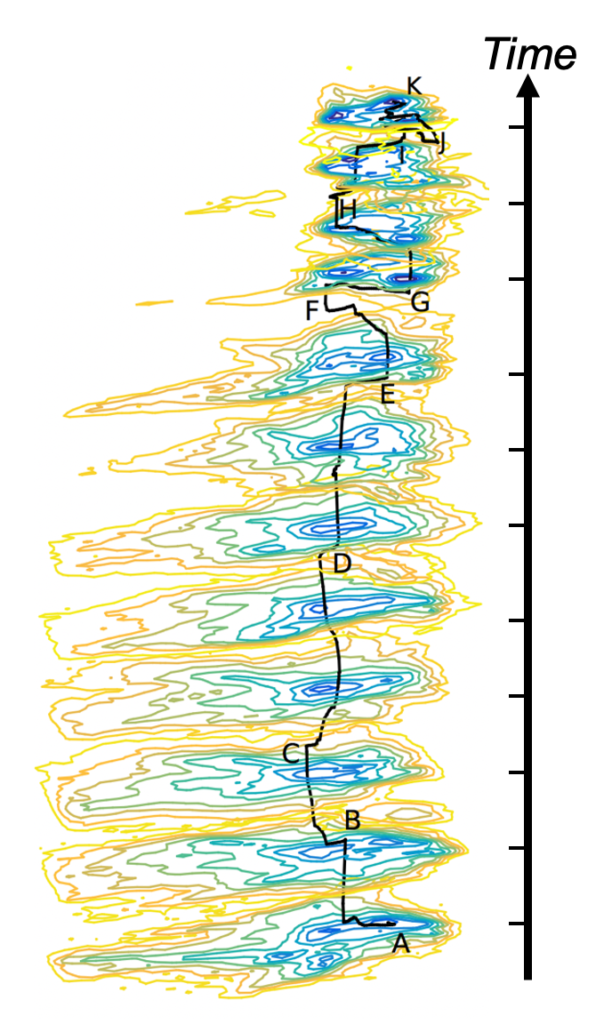
Recent progress in data analysis of single-particle cryogenic Electron Microscopy (cryoEM); such as Geometric Machine Learning algorithms, allows one to retrieve the conformational spectrum of heterogeneous molecular ensembles. Until recently, most efforts have been attempted under equilibrium conditions [1,2], where the conformational spectrum is time-independent, and thus directly yields the free-energy landscape. However, most biological systems operate far from equilibrium to sustain the processes that constitute life.
Under nonequilibrium conditions, the functional pathways are time-dependent, and the evolution of the conformational spectrum can be described by a Fokker-Planck equation, however with an unknown operator. State-of-the-art developments in Machine Learning, the so-called Physics-Informed Neural Networks (PINN) [3], allows one to retrieve the underlying Fokker-Planck operator from sparse observations alone [4], providing a complete physics-based description of the nonequilibrium process. Moreover, time-resolved cryoEM experiments have recently become practical [5]. Together, this enables us to combine the advantages of time-resolved serial crystallography (‘nonequilibrium processes’) with the advantages of single-particle methods (‘avoids averaging over unlike particles’).
Based on these opportunities, we present a conceptional and algorithmic framework to extract functional pathways from nonequilibrium from a collection of time-resolved single-particle images. This constitutes an unexplored route in studying biological function and structural dynamics under nonequilibrium conditions.
References
1. Dashti, A. et al. Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. U. S. A. 111, 17492–7 (2014).
2. Dashti, A. et al. Retrieving functional pathways of biomolecules from single-particle snapshots. Nat. Commun. 11, 4734 (2020).
3. Raissi, M., Perdikaris, P. & Karniadakis, G. E. Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J. Comput. Phys. 378, (2019).
4. Chen, X., Yang, L., Duan, J. & Karniadakis, G. E. Solving inverse stochastic problems from discrete particle observations using the Fokker-Planck equation and physics-informed neural networks. SIAM Journal on Scientific Computing 43, (2021).
5. Dandey, V. et al. Time-resolved cryo-EM using Spotiton, Nature Meth., 17, 897 (2020).